Staphylococcus aureus
2019-04-11
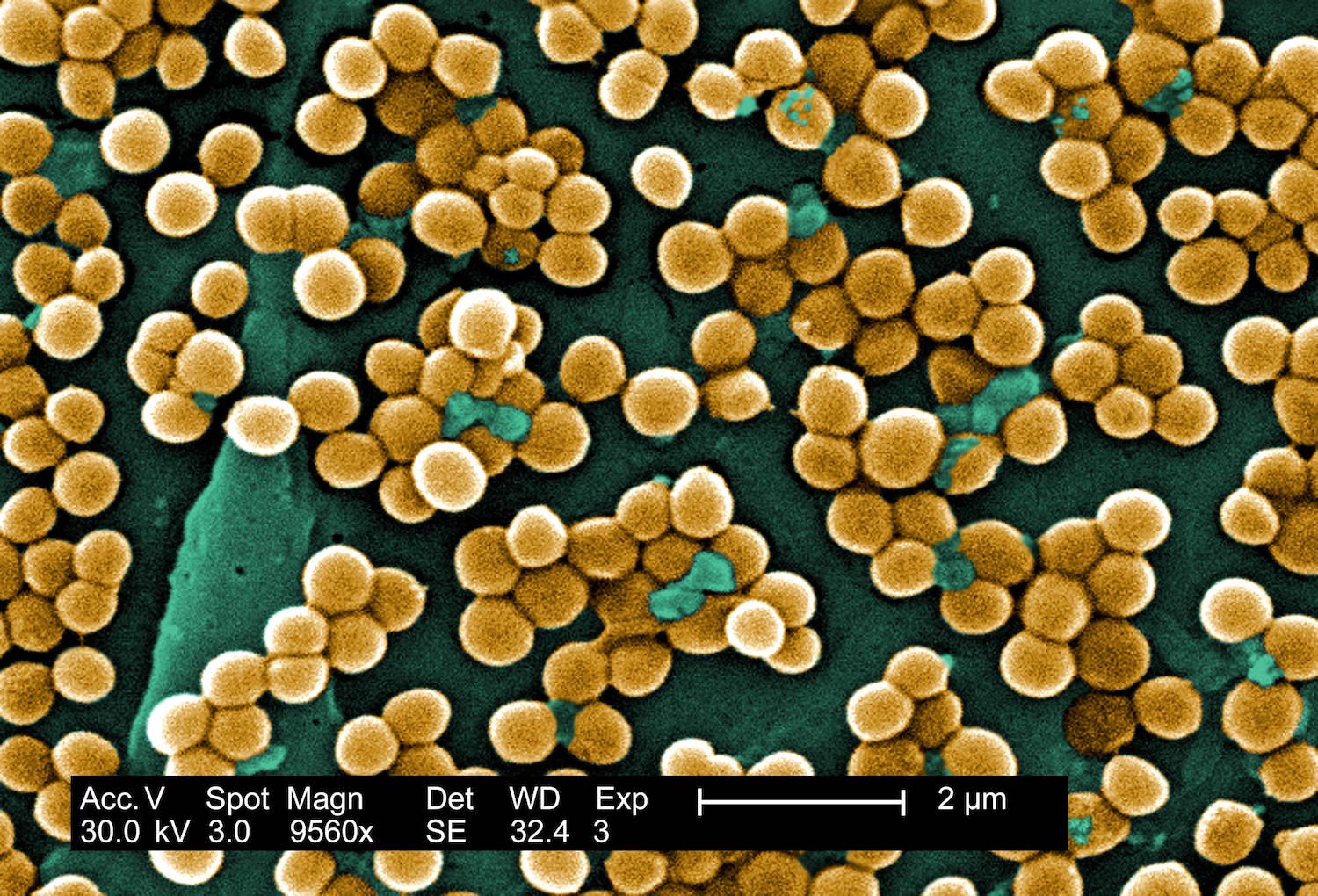
Digitally colorized scanning electron microscopic (SEM) of Staphylococcus aureus bacteria. Copyright holder: Janice Haney Carr. Link: https://phil.cdc.gov/Details.aspx?pid=10046.
S. aureus is a bacteria that normally lives on the skin or in the upper respiratory system. Almost a third of people in the United States have S. aureus living in their nostrils. Usually, it does not lead to any problems, but it has the power to cause opportunistic infections in most parts of the body and produce dangerous toxins. The most serious cases involve the bacteria infecting the bloodstream.
Before the discovery of antibiotics, 80% of people with S. aureus in the blood died. The antibiotic methicillin was discovered in 1960 that could effectively cure S. aureus infections. However, methicillin-resistant Staphylococcus aureus (MRSA) appeared in the clinic around one year or so after. This pathogen can also take up many other resistance genes from other bacteria to become even more dangerous and difficult to treat. The mortality rate continues to be far too high for bloodstream infections, ranging between 15% and 50%. Today, S. aureus is one of the most important pathogens responsible for hospital-acquired infections in wounds, after surgery and in the blood. In a hospital setting, virtually any object that comes into contact with the skin can pick up S. aureus and spread it around to other people and infect them.
It is more important now than ever before to get S. aureus (and especially MRSA) infections under control.
1928 offers the following analyses of S. aureus:
- MLST (Multilocus sequence typing).
- cgMLST (core genome MLST), typing with finer resolution than traditional MLST with our own schema built from 1704 genes.
- Staphylococcal cassette chromosome mec (SCCmec) typing.
- Virulence factors such as exfoliative toxins, leukocidins and superantigens.
- Genotypic prediction of resistance genes and mutations.
- SNP analysis for higher resolution of outbreaks.
Links and References
https://www.ncbi.nlm.nih.gov/pubmed/30737488