Klebsiella pneumoniae
2019-04-11
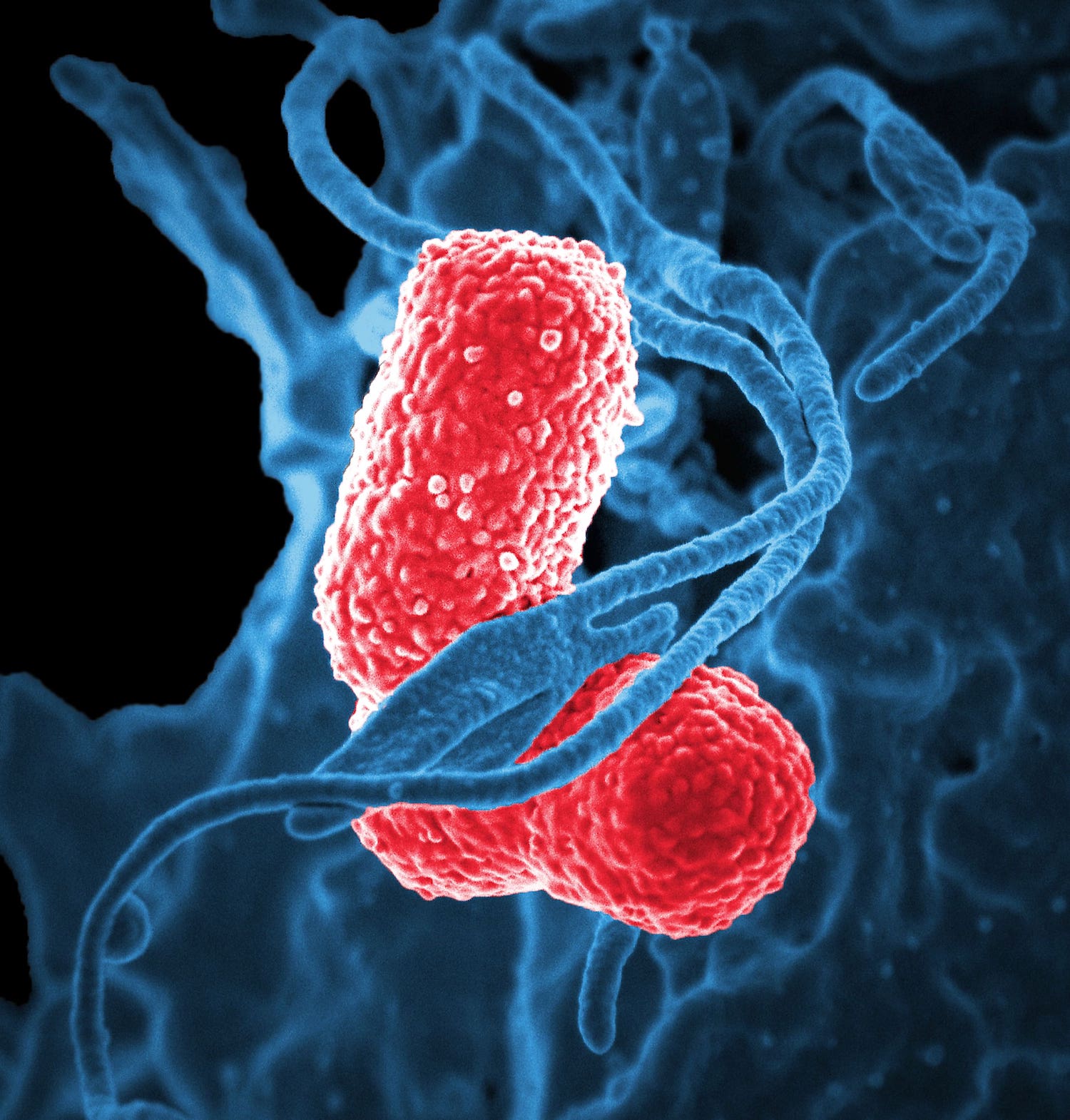
Digitally colorized scanning electron microscopic (SEM) of Klebsiella pneumoniae bacteria. Copyright holder: David Dorward; Ph.D.; National Institute of Allergy and Infectious Diseases (NIAID). Link: https://phil.cdc.gov/Details.aspx?pid=18170.
Increasing numbers of hospital acquired K. pneumoniae infections have obtained resistance to antibiotics through mutations and plasmids and many are CREs (Carbapenem Resistant Enterobacteriaceae). Carbapenems are often used as a last line antibiotic and the increased difficulty of treatment can increase the lethality of K. pneumoniae infections by up to 50% in infected patients according to the CDC.
This increase in resistance to last line antibiotics has placed K. pneumoniae on the WHO list of pathogens prioritized for R&D of new antibiotics.
Commonly found in the gut, these bacteria can be causative of infections such as pneumonia, bloodstream infection and meningitis if given the opportunity. In hospitals K. pneumoniae spread through person to person contact or through machinery like ventilators or intravenous catheters. 1928 has developed a pipeline for K. pneumoniae to track it and predict potential resistance genes.
1928 offers the following analyses of K. pneumoniae:
- MLST (Multilocus sequence typing).
- cgMLST (core genome MLST), typing with finer resolution than traditional MLST with our own schema built from 3459 genes.
- Genotypic prediction of resistance genes.
- SNP analysis for higher resolution of outbreaks.